Hyperparameter Tuning XGBoost with early stopping
This is a quick tutorial on how to tune the hyperparameters of an XGBoost model with a randomized search. It will also include early stopping to prevent overfitting and speed up training time. I believe the addition of early stopping helps set this apart from other tutorials on the topic.
Quick note that this post is almost identical to my post on hyperparameter tuning LightGBM with early stopping. It was surprisingly well received and has gotten more views than any of my other posts, so I decided to re-adapt it for XGBoost.
Why early stopping?
Early stopping is great. It helps prevent overfitting and it reduces the computational cost of training. It’s rare to get both of those at once - usually gains to model performance come at the cost of training time (e.g. lower learning rates + more epochs), and vice versa. One of my major takeaways from the acclaimed Deep Learning book is to always use early stopping if possible.
Like neural networks, gradient boosted trees are notorious for overfitting. Early stopping helps prevent this by stopping the training process when the model’s performance on a validation set stops improving. This is done by setting a threshold for the number of iterations the model can go without improving on the validation set. If the model fails to improve on the validation set for a number of iterations, the training process is stopped.
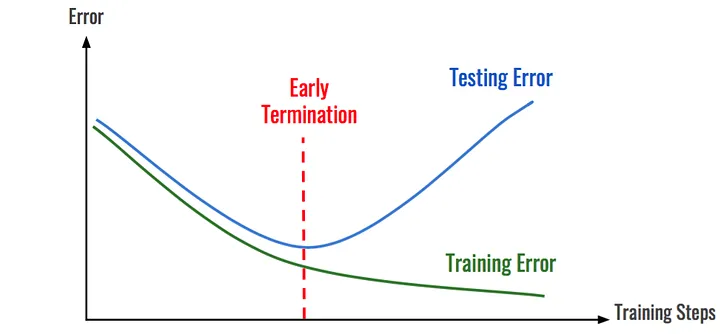
Computational cost is usually the limiting factor of hyperparameter tuning, so we want to decrease it as much as possible. In the mock example above, the training time was cut in half due to early stopping. This is a dramatic decrease, but the reduction in training time quickly becomes apparent when training hundreds or thousands of models while hyperparameter tuning.
Why a randomized search and not grid search?
Between grid search and random search, grid search generally makes more intuitive sense. However, research from James Bergstra and Yoshua Bengio have shown that random search tends to converge to good hyperparameters faster than grid search.
Here’s a graphic from their paper that gives an intuitive example of how random search can potentially cover more ground when there are hyperparameters that aren’t as important:

Source: James Bergstra & Yoshua Bengio
Additionally, grid search performs an exhaustive search over all possible combinations of hyperparameters, which can be prohibitively expensive to perform. Random search, on the other hand, only samples a fixed number of hyperparameter combinations, which can be set by the user. This allows the user to determine how much computational cost they would like to incur.
A quick note on hyperparameter tuning
Hyperparameter tuning helps improve the performance of a model, but it’s important to remember that it’s not the only thing that matters. Obviously it depends on the problem, but data cleaning and feature engineering are often more important to model performance than hyperparameter tuning. Collecting additional data (if possible) may also be more beneficial than hyperparameter tuning and can be checked with a learning curve.
The example
This example uses synthetic data to create a regresion problem. We’ll train a baseline model with the default parameters before performing hyperparameter tuning to see how we can improve on it.
import scipy
import numpy as np
import xgboost as xgb
from sklearn.model_selection import train_test_split
from sklearn.model_selection import RandomizedSearchCV
from sklearn.datasets import make_regression
from sklearn.metrics import mean_squared_error
# Create the dataset
X, y = make_regression(n_samples=50000, n_features=10, n_informative=5, noise=50, random_state=46)
# Split the data into train and test
X_train, X_test, y_train, y_test = train_test_split(X, y)
# Further splitting the training set into train and validation set for hyperparam tuning
X_train, X_val, y_train, y_val = train_test_split(X_train, y_train)
# Training a baseline model with the default parameters to see if we can improve on the results
baseline_model = xgb.XGBRegressor(n_jobs=-1, random_state=46)
baseline_model.fit(X_train, y_train)
print("Baseline model -")
print("R^2: ", baseline_model.score(X_test, y_test))
print("MSE: ", mean_squared_error(y_test, baseline_model.predict(X_test)))
Baseline model -
R^2: 0.9028353302352988
MSE: 3152.327724040228
Early stopping
To give a quick example of why you should almost always use early stopping with gradient boosting, here is the baseline model trained with early stopping. It is using the default 100 trees, but will stop if the performance on the validation set does not improve within 5 rounds. We’ll then compare the differences in training time and the model performance on the test set.
import time
start_time = time.time()
baseline_model.fit(X_train, y_train)
print("Training time for baseline model: ", time.time() - start_time)
early_stopping_model = xgb.XGBRegressor(early_stopping_rounds=5, n_jobs=-1, random_state=46)
start_time = time.time()
early_stopping_model.fit(X_train, y_train, eval_set=[(X_val, y_val)], verbose=False)
print("Training time for early stopping model: ", time.time() - start_time)
print("Early stopping model -")
print("Stopped on iteration: ", early_stopping_model.best_iteration)
print("R^2: ", early_stopping_model.score(X_test, y_test))
print("MSE: ", mean_squared_error(y_test, early_stopping_model.predict(X_test)))
Training time for baseline model: 0.14964723587036133
Training time for early stopping model: 0.10052227973937988
Early stopping model -
Stopped on iteration: 30
R^2: 0.9051606962683705
MSE: 3076.8855305727375
The model with early stopping finished training 1.5x faster and reduced the MSE by 76. The best iteration was 30, so the baseline model continued to train on an additional 70 trees that caused the model to begin overfitting.
Hyperparameter tuning
There are a handful of hyperparameter guides for XGBoost out there, but for this purpose we’ll borrow from this guide written by Jason Brownlee from Machine Learning Mastery and mix in a few of the parameters from Leonie Monigatti’s LightGBM hyperparameter tuning guide. These are just suggestions, and I recommend reading about the parameters from the documentation to better understand how changing them affects the model.

Credit: Leonie Monigatti
I mentioned above that we can set the number of iterations for the random search. I’m setting it to 40 to keep the runtime relatively short, but you can increase it to be more likely to get better results. I am also using 10,000 trees and 20 rounds before early stopping, these were chosen somewhat arbitrarily. Ideally early stopping will prevent most runs from training the full 10,000 trees, but more trees will likely be trained if the learning rate is lower which would result in a higher computational cost. A higher number of early stopping rounds will help ensure that the model does not stop prematurely, but it will increase the computational cost by continuing to train additional trees after it is likely beginning to overfit.
Note that I am using k-folds cross validation here, but I included code for using a train/validation/test split. Just uncomment it and remove the train/test split portions.
Option 1: Using scikit-learn’s RandomizedSearchCV()
This option looks ideal at first glance because it’s using a function one of the best machine learning libraries. However, something a little goofy is happening. The cv parameter of the RandomizedSearchCV() function is further splitting the training set and using the newly split part as the validation set, whereas the eval_set argument in the fit() method is using the pre-determined validation set.
This effectively results in two different validation sets being used for two different things - one for early stopping, and one to select the hyperparameters. Not only could this result in the model being less likely to find the optimal hyparparameters because it is validating on two different data sets, but it also uses a smaller amount of training data. The XGBoost documentation recommends re-training with early stopping after performing hyperparameter tuning with cross-validation since the number of trees could differ for each fold, so we will do this at the end.
Despite these issues, this still trains faster than had early stopping not been used and makes the model less likely to overfit. We’ll run this, see how the performance compares to the non-tuned early stopping model, and then we’ll evaluate another option that uses a custom built version of the randomized search with cross-validation.
# Define a new model object with a large number of estimators since we will be using early stopping
# The early stopping rounds must be called in the model initialization to work with RandomizedSearchCV()
model = xgb.XGBRegressor(n_estimators=10000, early_stopping_rounds=20, n_jobs=-1, random_state=46)
# Define the parameter distributions for hyperparameter tuning
# Using this guide: https://machinelearningmastery.com/configure-gradient-boosting-algorithm/
# Parameter documentation: https://xgboost.readthedocs.io/en/stable/parameter.html
param_distributions = {
"learning_rate": scipy.stats.uniform(loc=0.003, scale=0.19), # Default is 0.3. Ranges from loc to loc+scale.
"subsample": scipy.stats.uniform(loc=0.5, scale=0.5), # Default is 1
"colsample_bytree": scipy.stats.uniform(loc=0.5, scale=0.5), # Default is 1
"min_child_weight": [1, 3, 5, 7], # Default is 1
"max_depth": np.append(0, np.arange(3, 16)), # Default is 6
"alpha": [0, 0.01, 1, 2, 5, 7, 10, 50, 100], # Default is 0. AKA reg_alpha.
"lambda": [0, 0.01, 1, 5, 10, 20, 50, 100] # Default is 0. AKA reg_lambda.
}
# Configure the randomized search
random_search = RandomizedSearchCV(model,
param_distributions=param_distributions,
n_iter=40,
cv=3, # k-folds
# cv=ShuffleSplit(n_splits=1, test_size=.2, random_state=46), # Train/test split
scoring="neg_mean_squared_error",
n_jobs=-1)
# Perform the randomized search with early stopping
random_search.fit(X_train, y_train,
eval_set=[(X_val, y_val)],
verbose=False)
# Extract the tuned model from the random search
tuned_model = random_search.best_estimator_
print("Tuned model -")
print("R^2: ", tuned_model.score(X_test, y_test))
print("MSE: ", mean_squared_error(y_test, tuned_model.predict(X_test)), "\n")
# The XGBoost documentation recommends re-training the model with early stopping after the optimal hyperparameters are found with cross validation
# This is because the number of trees will likely change with each fold
# https://xgboost.readthedocs.io/en/stable/python/sklearn_estimator.html#early-stopping
optimal_params = tuned_model.get_params()
optimal_params["n_estimators"] = tuned_model.best_iteration
tuned_retrained_model = xgb.XGBRegressor(**optimal_params) # Inherits n_jobs and random_state from above
tuned_retrained_model.fit(X_train, y_train, eval_set=[(X_val, y_val)], verbose=False)
print("Tuned re-trained model -")
print("R^2: ", tuned_retrained_model.score(X_test, y_test))
print("MSE: ", mean_squared_error(y_test, tuned_retrained_model.predict(X_test)))
Tuned model -
R^2: 0.9185246168223384
MSE: 2643.317883338798
Tuned re-trained model -
R^2: 0.9185245088513797
MSE: 2643.3213862565526
Compared to the non-tuned early stopping model we were able to improve our R^2 score by about 1.5% and reduce our MSE by about 14% by tuning the hyperparameters. Let’s see what the optimal parameters were:
# Show the optimal parameters that were obtained
{key: value for key, value in optimal_params.items() if value is not None}
{'objective': 'reg:squarederror',
'colsample_bytree': 0.6829993834296832,
'early_stopping_rounds': 20,
'enable_categorical': False,
'learning_rate': 0.04469805257577043,
'max_depth': 3,
'min_child_weight': 1,
'missing': nan,
'n_estimators': 690,
'n_jobs': -1,
'random_state': 46,
'subsample': 0.6672594763976536,
'alpha': 5,
'lambda': 100}
Option 2: Manually
This option involves more code and a handful of for loops that are not exactly optimized. However, it will result in ensuring that the same validation set is being used both for the early stopping and for the hyperparameter search.
Note that this includes options for both a train/validation/test split and using k-folds cross validation that is being determined by the tune_with_kfolds parameter. This could be much shorter if only using one, but I wanted to include both options.
from sklearn.base import clone
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
from tqdm import tqdm
# Use train/val/test split or k-folds cross validation
tune_with_kfolds = True
# Define the parameter distributions for hyperparameter tuning
# Using this guide: https://machinelearningmastery.com/configure-gradient-boosting-algorithm/
# Parameter documentation: https://xgboost.readthedocs.io/en/stable/parameter.html
param_distributions = {
"learning_rate": scipy.stats.uniform(loc=0.003, scale=0.19), # Default is 0.3. Ranges from loc to loc+scale.
"subsample": scipy.stats.uniform(loc=0.5, scale=0.5), # Default is 1
"colsample_bytree": scipy.stats.uniform(loc=0.5, scale=0.5), # Default is 1
"min_child_weight": [1, 3, 5, 7], # Default is 1
"max_depth": np.append(0, np.arange(3, 16)), # Default is 6
"alpha": [0, 0.01, 1, 2, 5, 7, 10, 50, 100], # Default is 0. AKA reg_alpha.
"lambda": [0, 0.01, 1, 5, 10, 20, 50, 100] # Default is 0. AKA reg_lambda.
}
def sample_from_param_distributions(param_distributions: dict) -> dict:
"""
Sample a value from each parameter distribution defined in param_distributions.
Parameters:
- param_distributions (dict): Dictionary where keys are parameter names and values are either:
- scipy.stats distribution objects for continuous distributions.
- Lists or numpy arrays for discrete choices.
Returns:
- sampled_values (dict): Dictionary containing sampled values corresponding to each parameter.
"""
sampled_values = {}
for param, distribution in param_distributions.items():
if isinstance(distribution, scipy.stats._distn_infrastructure.rv_frozen):
sampled_values[param] = distribution.rvs()
elif isinstance(distribution, list) or isinstance(distribution, np.ndarray):
sampled_values[param] = np.random.choice(distribution)
else:
raise ValueError(f"Unsupported distribution type for parameter '{param}'")
return sampled_values
num_iterations = 40
optimal_params = {}
best_score = -np.inf
for iteration in tqdm(range(num_iterations)):
# Sample values from the distributions
sampled_params = sample_from_param_distributions(param_distributions)
# Train the model, get the performance on the validation set
model = xgb.XGBRegressor(n_estimators=10000, early_stopping_rounds=20,
n_jobs=-1, random_state=46, **sampled_params)
# Perform the tuning with either k-folds or train/test split
if tune_with_kfolds == True:
cv = KFold(n_splits=3, shuffle=True, random_state=46) # Use StratifiedKFold for classification
cv_results = []
for train_idx, val_idx in cv.split(X_train, y_train):
X_train_fold, X_val_fold = X_train[train_idx], X_train[val_idx]
y_train_fold, y_val_fold = y_train[train_idx], y_train[val_idx]
model_clone = clone(model)
model_clone.fit(X_train_fold, y_train_fold, eval_set=[(X_val_fold, y_val_fold)], verbose=False)
predictions = model_clone.predict(X_val_fold)
fold_neg_mse = -mean_squared_error(y_true=y_val_fold, y_pred=predictions)
cv_results.append(fold_neg_mse)
neg_mean_squared_error = np.mean(cv_results)
else:
# Train/test split with the validation data set for early stopping
model.fit(X_train, y_train, eval_set=[(X_val, y_val)], verbose=False)
predictions = model.predict(X_val)
neg_mean_squared_error = -mean_squared_error(y_true=y_val, y_pred=predictions)
# Set the optimal parameters if the performance is better
if neg_mean_squared_error > best_score:
best_score = neg_mean_squared_error
optimal_params = sampled_params
# Need to re-train w/ early stopping to get optimal number of estimators if tuned with k-folds
if tune_with_kfolds == False:
optimal_params["n_estimators"] = model.best_iteration
# Re-train with the optimal hyperparams
# Re-perform early stopping if k-folds was used for tuning
if tune_with_kfolds == True:
tuned_model = xgb.XGBRegressor(**optimal_params, n_jobs=-1, random_state=46,
n_estimators=10000, early_stopping_rounds=20)
tuned_model.fit(X_train, y_train, eval_set=[(X_val, y_val)], verbose=False)
optimal_params["n_estimators"] = tuned_model.best_iteration
else:
tuned_model = xgb.XGBRegressor(**optimal_params, n_jobs=-1, random_state=46)
tuned_model.fit(X_train, y_train)
# Report the results
print("Tuned model -")
print("R^2: ", tuned_model.score(X_test, y_test))
print("MSE: ", mean_squared_error(y_test, tuned_model.predict(X_test)))
100%|██████████| 40/40 [05:58<00:00, 8.97s/it]
Tuned model -
R^2: 0.9191275594609924
MSE: 2623.756526310182
This option seemingly performed better than the RandomizedSearchCV() option, but it could be due to the randomness. This custom implementation is less optimized (especially for k-folds) so it will likely take longer to run than the first option.
Next steps
There are ways to continue taking this further, but I want to keep this post relatively short. The next steps I would take would be to increase the number of iterations for the random search, and then potentially use a lower learning rate to fine-tune the model even further. There are stepwise strategies for hyperparameter tuning an XGBoost model that could also be tried. Depending on the problem, there are likely different things that can be done with the data to improve the model as well.
If you have any questions or comments, please feel free to reach out to me on by email or on LinkedIn. Thanks for reading!